| 1 | #!/usr/local/bin/python |
|---|
| 2 | |
|---|
| 3 | import sys |
|---|
| 4 | import re |
|---|
| 5 | import os |
|---|
| 6 | import random |
|---|
| 7 | import copy |
|---|
| 8 | |
|---|
| 9 | from optparse import OptionParser, OptionValueError |
|---|
| 10 | from federation.remote_service import service_caller |
|---|
| 11 | from federation.service_error import service_error |
|---|
| 12 | from federation import topdl |
|---|
| 13 | |
|---|
| 14 | class constraint: |
|---|
| 15 | """ |
|---|
| 16 | This is just a struct to hold constraint fields, really |
|---|
| 17 | """ |
|---|
| 18 | def __init__(self, name=None, required=False, accepts=None, provides=None, |
|---|
| 19 | match=None): |
|---|
| 20 | self.name = name |
|---|
| 21 | self.required = required |
|---|
| 22 | self.accepts = accepts or [] |
|---|
| 23 | self.provides = provides or [] |
|---|
| 24 | self.match = match |
|---|
| 25 | |
|---|
| 26 | def __str__(self): |
|---|
| 27 | return "%s:%s:%s:%s" % (self.name, self.required, |
|---|
| 28 | ",".join(self.provides), ",".join(self.accepts)) |
|---|
| 29 | |
|---|
| 30 | |
|---|
| 31 | class ComposeOptionParser(OptionParser): |
|---|
| 32 | """ |
|---|
| 33 | This class encapsulates the options to this script in one place. It also |
|---|
| 34 | holds the callback for the multifile choice. |
|---|
| 35 | """ |
|---|
| 36 | def __init__(self): |
|---|
| 37 | OptionParser.__init__(self) |
|---|
| 38 | self.add_option('--url', dest='url', default="http://localhost:13235", |
|---|
| 39 | help='url of ns2 to topdl service') |
|---|
| 40 | self.add_option('--certfile', dest='cert', default=None, |
|---|
| 41 | help='Certificate to use as identity') |
|---|
| 42 | self.add_option('--seed', dest='seed', type='int', default=None, |
|---|
| 43 | help='Random number seed') |
|---|
| 44 | self.add_option('--multifile', dest='files', default=[], type='string', |
|---|
| 45 | action='callback', callback=self.multi_callback, |
|---|
| 46 | help="Include file multiple times") |
|---|
| 47 | self.add_option('--output', dest='outfile', default=None, |
|---|
| 48 | help='Output file name') |
|---|
| 49 | self.add_option('--format', dest='format', default="xml", |
|---|
| 50 | help='Output file format') |
|---|
| 51 | self.add_option('--add_testbeds', dest='add_testbeds', default=False, |
|---|
| 52 | action='store_true', |
|---|
| 53 | help='add testbed attributes to each component') |
|---|
| 54 | self.add_option('--lax', dest='lax', default=False, |
|---|
| 55 | action='store_true', |
|---|
| 56 | help='allow solutions where unrequired constraints fail') |
|---|
| 57 | |
|---|
| 58 | @staticmethod |
|---|
| 59 | def multi_callback(option, opt_str, value, parser): |
|---|
| 60 | """ |
|---|
| 61 | Parse a --multifile command line option. The parameter is of the form |
|---|
| 62 | filename,count. This splits the argument at the rightmost comma and |
|---|
| 63 | inserts the filename, count tuple into the "files" option. It handles |
|---|
| 64 | a couple error cases, too. |
|---|
| 65 | """ |
|---|
| 66 | idx = value.rfind(',') |
|---|
| 67 | if idx != -1: |
|---|
| 68 | try: |
|---|
| 69 | parser.values.files.append((value[0:idx], int(value[idx+1:]))) |
|---|
| 70 | except ValueError, e: |
|---|
| 71 | raise OptionValueError( |
|---|
| 72 | "Can't convert %s to int in multifile (%s)" % \ |
|---|
| 73 | (value[idx+1:], value)) |
|---|
| 74 | else: |
|---|
| 75 | raise OptionValueError( |
|---|
| 76 | "Bad format (need a comma) for multifile: %s" % value) |
|---|
| 77 | |
|---|
| 78 | def warn(msg): |
|---|
| 79 | """ |
|---|
| 80 | Exactly what you think. Print a message to stderr |
|---|
| 81 | """ |
|---|
| 82 | print >>sys.stderr, msg |
|---|
| 83 | |
|---|
| 84 | |
|---|
| 85 | def make_new_name(names, prefix="name"): |
|---|
| 86 | """ |
|---|
| 87 | Generate an identifier not present in names by appending an integer to |
|---|
| 88 | prefix. The new name is inserted into names and returned. |
|---|
| 89 | """ |
|---|
| 90 | i = 0 |
|---|
| 91 | n = "%s%d" % (prefix,i) |
|---|
| 92 | while n in names: |
|---|
| 93 | i += 1 |
|---|
| 94 | n = "%s%d" % (prefix,i) |
|---|
| 95 | names.add(n) |
|---|
| 96 | return n |
|---|
| 97 | |
|---|
| 98 | def add_interfaces(top, mark): |
|---|
| 99 | """ |
|---|
| 100 | Add unconnected interfaces to the nodes whose names are keys in the mark |
|---|
| 101 | dict. As the interface is added change the name field of the contstraint |
|---|
| 102 | in mark into a tuple containing the node name, interface name, and topology |
|---|
| 103 | in which they reside. |
|---|
| 104 | """ |
|---|
| 105 | for e in top.elements: |
|---|
| 106 | if e.name in mark: |
|---|
| 107 | con = mark[e.name] |
|---|
| 108 | ii = make_new_name(set([x.name for x in e.interface]), "inf") |
|---|
| 109 | e.interface.append( |
|---|
| 110 | topdl.Interface(substrate=[], name=ii, element=e)) |
|---|
| 111 | con.name = (e.name, ii, top) |
|---|
| 112 | |
|---|
| 113 | def localize_names(top, names, marks): |
|---|
| 114 | """ |
|---|
| 115 | Take a topology and rename any substrates or elements that share a name |
|---|
| 116 | with an existing computer or substrate. Keep the original name as a |
|---|
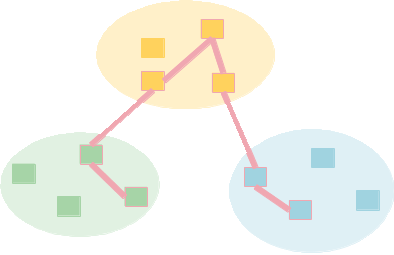
| 117 | localized_name attribute. In addition, remap any constraints or interfaces |
|---|
| 118 | that point to the old name over to the new one. Constraints are found in |
|---|
| 119 | the marks dict, indexed by node name. Those constraints name attributes |
|---|
| 120 | have already been converted to triples (node name, interface name, |
|---|
| 121 | topology) so only the node name needs to be changed. |
|---|
| 122 | """ |
|---|
| 123 | sub_map = { } |
|---|
| 124 | for s in top.substrates: |
|---|
| 125 | s.set_attribute('localized_name', s.name) |
|---|
| 126 | if s.name in names: |
|---|
| 127 | sub_map[s.name] = n = make_new_name(names, "substrate") |
|---|
| 128 | s.name = n |
|---|
| 129 | else: |
|---|
| 130 | names.add(s.name) |
|---|
| 131 | |
|---|
| 132 | for e in [ e for e in top.elements if isinstance(e, topdl.Computer)]: |
|---|
| 133 | e.set_attribute('localized_name', e.name) |
|---|
| 134 | if e.name in names: |
|---|
| 135 | nn= make_new_name(names, "computer") |
|---|
| 136 | if e.name in marks: |
|---|
| 137 | n, i, t = marks[e.name].name |
|---|
| 138 | marks[e.name].name = (nn, i, t) |
|---|
| 139 | e.name = nn |
|---|
| 140 | else: |
|---|
| 141 | names.add(e.name) |
|---|
| 142 | |
|---|
| 143 | # Interface mapping. The list comprehension creates a list of |
|---|
| 144 | # substrate names where each element in the list is replaced by the |
|---|
| 145 | # entry in sub_map indexed by it if present and left alone otherwise. |
|---|
| 146 | for i in e.interface: |
|---|
| 147 | i.substrate = [ sub_map.get(ii, ii) for ii in i.substrate ] |
|---|
| 148 | |
|---|
| 149 | def meet_constraints(candidates, provides, accepts): |
|---|
| 150 | """ |
|---|
| 151 | Try to meet all the constraints in candidates using the information in the |
|---|
| 152 | provides and accepts dicts (which index constraints that provide or accept |
|---|
| 153 | the given attribute). A constraint is met if it can be matched with another |
|---|
| 154 | constraint that provides an attribute that the first constraint accepts. |
|---|
| 155 | Only one match per pair is allowed, and we always prefer matching a |
|---|
| 156 | required constraint to an unreqired one. If all the candidates can be |
|---|
| 157 | matches, return True and return False otherwise. |
|---|
| 158 | """ |
|---|
| 159 | got_all = True |
|---|
| 160 | for c in candidates: |
|---|
| 161 | if not c.match: |
|---|
| 162 | rmatch = None # Match to a required constraint |
|---|
| 163 | umatch = None # Match to an unrequired constraint |
|---|
| 164 | for a in c.accepts: |
|---|
| 165 | for can in provides.get(a,[]): |
|---|
| 166 | # A constraint cannot satisfy itself, nor do we allow loops |
|---|
| 167 | # within the same topology. This also excludes matched |
|---|
| 168 | # constraints. |
|---|
| 169 | if can.name != c.name and can.name[2] != c.name[2] and \ |
|---|
| 170 | not can.match: |
|---|
| 171 | # Now check that the candidate also accepts c |
|---|
| 172 | for ca in can.accepts: |
|---|
| 173 | if ca in c.provides: |
|---|
| 174 | if can.required: rmatch = can |
|---|
| 175 | else: umatch = can |
|---|
| 176 | break |
|---|
| 177 | |
|---|
| 178 | # Within providers, prefer matches against required |
|---|
| 179 | # composition points. |
|---|
| 180 | if rmatch: |
|---|
| 181 | break |
|---|
| 182 | # Within acceptance categories, prefer matches against required |
|---|
| 183 | # composition points |
|---|
| 184 | if rmatch: |
|---|
| 185 | break |
|---|
| 186 | |
|---|
| 187 | # Move the better match over to the match variable |
|---|
| 188 | if rmatch: match = rmatch |
|---|
| 189 | elif umatch: match = umatch |
|---|
| 190 | else: match = None |
|---|
| 191 | |
|---|
| 192 | # Done checking all possible matches. Record the match or note an |
|---|
| 193 | # unmatched candidate. |
|---|
| 194 | if match: |
|---|
| 195 | match.match = c |
|---|
| 196 | c.match = match |
|---|
| 197 | else: |
|---|
| 198 | got_all = False |
|---|
| 199 | return got_all |
|---|
| 200 | |
|---|
| 201 | def randomize_constraint_order(indexes): |
|---|
| 202 | """ |
|---|
| 203 | Randomly reorder the lists of constraints that provides and accepts hold. |
|---|
| 204 | """ |
|---|
| 205 | if not isinstance(indexes, tuple): |
|---|
| 206 | indexes = (indexes,) |
|---|
| 207 | |
|---|
| 208 | for idx in indexes: |
|---|
| 209 | for k in idx.keys(): |
|---|
| 210 | random.shuffle(idx[k]) |
|---|
| 211 | |
|---|
| 212 | def remote_ns2topdl(uri, desc, cert): |
|---|
| 213 | """ |
|---|
| 214 | Call a remote service to convert the ns2 to topdl (and in fact all the way |
|---|
| 215 | to a topdl.Topology. |
|---|
| 216 | """ |
|---|
| 217 | |
|---|
| 218 | req = { 'description' : { 'ns2description': desc }, } |
|---|
| 219 | |
|---|
| 220 | r = service_caller('Ns2Topdl')(uri, req, cert) |
|---|
| 221 | |
|---|
| 222 | if r.has_key('Ns2TopdlResponseBody'): |
|---|
| 223 | r = r['Ns2TopdlResponseBody'] |
|---|
| 224 | ed = r.get('experimentdescription', None) |
|---|
| 225 | if 'topdldescription' in ed: |
|---|
| 226 | return topdl.Topology(**ed['topdldescription']) |
|---|
| 227 | else: |
|---|
| 228 | return None |
|---|
| 229 | else: |
|---|
| 230 | return None |
|---|
| 231 | |
|---|
| 232 | def connect_composition_points(top, contraints, names): |
|---|
| 233 | """ |
|---|
| 234 | top is a topology containing copies of all the topologies represented in |
|---|
| 235 | the contsraints, flattened into one name space. This routine inserts the |
|---|
| 236 | additional substrates required to interconnect the topology as described by |
|---|
| 237 | the constraints. After the links are made, unused connection points are |
|---|
| 238 | purged. |
|---|
| 239 | """ |
|---|
| 240 | done = set() |
|---|
| 241 | for c in constraints: |
|---|
| 242 | if c not in done and c.match: |
|---|
| 243 | # c is an unprocessed matched constraint. Create a substrate |
|---|
| 244 | # and attach it to the interfaces named by c and c.match. |
|---|
| 245 | sn = make_new_name(names, "sub") |
|---|
| 246 | s = topdl.Substrate(name=sn) |
|---|
| 247 | connected = 0 |
|---|
| 248 | # Walk through all the computers in the topology |
|---|
| 249 | for e in [ e for e in top.elements \ |
|---|
| 250 | if isinstance(e, topdl.Computer)]: |
|---|
| 251 | # if the computer matches the computer and interface name in |
|---|
| 252 | # either c or c.match, connect it. Once both computers have |
|---|
| 253 | # been found and connected, exit the loop walking through all |
|---|
| 254 | # computers. |
|---|
| 255 | for comp, inf in (c.name[0:2], c.match.name[0:2]): |
|---|
| 256 | if e.name == comp: |
|---|
| 257 | for i in e.interface: |
|---|
| 258 | if i.name == inf: |
|---|
| 259 | i.substrate.append(sn) |
|---|
| 260 | connected += 1 |
|---|
| 261 | break |
|---|
| 262 | break |
|---|
| 263 | # Connected both, so add the substrate to the topology |
|---|
| 264 | if connected == 2: |
|---|
| 265 | top.substrates.append(s) |
|---|
| 266 | break |
|---|
| 267 | # c and c.match have been processed, so add them to the done set |
|---|
| 268 | done.add(c) |
|---|
| 269 | done.add(c.match) |
|---|
| 270 | |
|---|
| 271 | # All interfaces with matched constraints have been connected. Cull any |
|---|
| 272 | # interfaces unassigned to a substrate |
|---|
| 273 | for e in [ e for e in top.elements if isinstance(e, topdl.Computer)]: |
|---|
| 274 | if any([ not i.substrate for i in e.interface]): |
|---|
| 275 | e.interface = [ i for i in e.interface if i.substrate ] |
|---|
| 276 | |
|---|
| 277 | top.incorporate_elements() |
|---|
| 278 | |
|---|
| 279 | def import_ns2_constraints(contents): |
|---|
| 280 | """ |
|---|
| 281 | Contents is a list containing the lines of an annotated ns2 file. This |
|---|
| 282 | routine extracts the constraint comment lines and convertes them into |
|---|
| 283 | constraints in the namespace of the tcl experiment, as well as inserting |
|---|
| 284 | them in the accepts and provides indices. |
|---|
| 285 | |
|---|
| 286 | Constraints are given in lines of the form |
|---|
| 287 | name:required:provides:accepts |
|---|
| 288 | where name is the name of the node in the current topology, if the second |
|---|
| 289 | field is "required" this is a required constraint, a list of attributes |
|---|
| 290 | that this connection point provides and a list that it accepts. The |
|---|
| 291 | provides and accepts lists are comma-separated. The constraint is added to |
|---|
| 292 | the marks dict under the name key and that dict is returned. |
|---|
| 293 | """ |
|---|
| 294 | |
|---|
| 295 | const_re = re.compile("\s*#\s*COMPOSITION:\s*([^:]+:[^:]+:.*)") |
|---|
| 296 | |
|---|
| 297 | marks = { } |
|---|
| 298 | for l in contents: |
|---|
| 299 | m = const_re.search(l) |
|---|
| 300 | if m: |
|---|
| 301 | exp = re.sub('\s', '', m.group(1)) |
|---|
| 302 | nn, r, p, a = exp.split(":") |
|---|
| 303 | if nn.find('(') != -1: |
|---|
| 304 | # Convert array references into topdl names |
|---|
| 305 | nn = re.sub('\\((\\d)\\)', '-\\1', nn) |
|---|
| 306 | p = p.split(",") |
|---|
| 307 | a = a.split(",") |
|---|
| 308 | c = constraint(name=nn, required=(r == 'required'), |
|---|
| 309 | provides=p, accepts=a) |
|---|
| 310 | marks[nn] = c |
|---|
| 311 | return marks |
|---|
| 312 | |
|---|
| 313 | def add_constraint_attributes(top, marks): |
|---|
| 314 | """ |
|---|
| 315 | Take the constraints in marks and add attributes representing them to the |
|---|
| 316 | nodes in top. |
|---|
| 317 | """ |
|---|
| 318 | for e in [e for e in top.elements if isinstance(e, topdl.Computer)]: |
|---|
| 319 | if e.name in marks and not e.get_attribute('composition_point'): |
|---|
| 320 | c = marks[e.name] |
|---|
| 321 | e.set_attribute('composition_point', 'true') |
|---|
| 322 | e.set_attribute('required', '%s' % c.required) |
|---|
| 323 | e.set_attribute('provides', ",".join(c.provides)) |
|---|
| 324 | e.set_attribute('accepts', ",".join(c.accepts)) |
|---|
| 325 | |
|---|
| 326 | def remove_constraint_attributes(top, marks): |
|---|
| 327 | """ |
|---|
| 328 | Remove constraint attributes that match the ones in the dict from the |
|---|
| 329 | topology. |
|---|
| 330 | """ |
|---|
| 331 | for e in [e for e in top.elements \ |
|---|
| 332 | if isinstance(e, topdl.Computer) and e.name in marks]: |
|---|
| 333 | e.remove_attribute('composition_point') |
|---|
| 334 | e.remove_attribute('required') |
|---|
| 335 | e.remove_attribute('provides') |
|---|
| 336 | e.remove_attribute('accepts') |
|---|
| 337 | |
|---|
| 338 | def import_ns2_component(fn): |
|---|
| 339 | """ |
|---|
| 340 | Pull a composition component in from an ns2 description. The Constraints |
|---|
| 341 | are parsed from the comments using import_ns2_constraints and the topology |
|---|
| 342 | is created using a call to a fedd exporting the Ns2Topdl function. A |
|---|
| 343 | topdl.Topology object rempresenting the component's topology and a dict |
|---|
| 344 | mapping constraints from the components names to the conttraints is |
|---|
| 345 | returned. If either the file read or the conversion fails, appropriate |
|---|
| 346 | Exceptions are thrown. |
|---|
| 347 | """ |
|---|
| 348 | f = open(fn, "r") |
|---|
| 349 | contents = [ l for l in f ] |
|---|
| 350 | |
|---|
| 351 | marks = import_ns2_constraints(contents) |
|---|
| 352 | top = remote_ns2topdl(opts.url, "".join(contents), cert) |
|---|
| 353 | if not top: |
|---|
| 354 | raise RuntimeError("Cannot create topology from: %s" % fn) |
|---|
| 355 | add_constraint_attributes(top, marks) |
|---|
| 356 | |
|---|
| 357 | return (top, marks) |
|---|
| 358 | |
|---|
| 359 | def import_topdl_component(fn): |
|---|
| 360 | """ |
|---|
| 361 | Pull a component in from a topdl description. The topology generation is |
|---|
| 362 | straightforward and the constraints are pulled from the attributes of |
|---|
| 363 | Computer nodes in the experiment. The compisition_point attribute being |
|---|
| 364 | true marks a node as a composition point. The required, provides and |
|---|
| 365 | accepts attributes map directly into those constraint fields. The dict of |
|---|
| 366 | constraints and the topology are returned. |
|---|
| 367 | """ |
|---|
| 368 | top = topdl.topology_from_xml(filename=fn, top='experiment') |
|---|
| 369 | marks = { } |
|---|
| 370 | for e in [ e for e in top.elements if isinstance(e, topdl.Computer)]: |
|---|
| 371 | if e.get_attribute('composition_point'): |
|---|
| 372 | r = e.get_attribute('required') or 'false' |
|---|
| 373 | a = e.get_attribute('accepts') |
|---|
| 374 | p = e.get_attribute('provides') |
|---|
| 375 | if a and p: |
|---|
| 376 | c = constraint(name=e.name, required=(r == 'required'), |
|---|
| 377 | provides=p.split(','), accepts=a.split(',')) |
|---|
| 378 | marks[e.name] = c |
|---|
| 379 | return (top, marks) |
|---|
| 380 | |
|---|
| 381 | def index_constraints(constraints, provides, accepts): |
|---|
| 382 | """ |
|---|
| 383 | Add constraints to the provides and accepts indices based on the attributes |
|---|
| 384 | of the contstraints. |
|---|
| 385 | """ |
|---|
| 386 | for c in constraints: |
|---|
| 387 | for attr, dict in ((c.provides, provides), (c.accepts, accepts)): |
|---|
| 388 | for a in attr: |
|---|
| 389 | if a not in dict: dict[a] = [c] |
|---|
| 390 | else: dict[a].append(c) |
|---|
| 391 | |
|---|
| 392 | def get_suffix(fn): |
|---|
| 393 | """ |
|---|
| 394 | We get filename suffixes a couple places. It;s worth using the same code. |
|---|
| 395 | This gets the shortest . separated suffix from a filename, or None. |
|---|
| 396 | """ |
|---|
| 397 | idx = fn.rfind('.') |
|---|
| 398 | if idx != -1: return fn[idx+1:] |
|---|
| 399 | else: return None |
|---|
| 400 | |
|---|
| 401 | |
|---|
| 402 | def output_composition(top, constraints, outfile=None, format=None): |
|---|
| 403 | """ |
|---|
| 404 | Output the composition to the file named by outfile (if any) in the format |
|---|
| 405 | given by format if any. If both are None, output to stdout in topdl |
|---|
| 406 | """ |
|---|
| 407 | def topdl_out(f, top, constraints): |
|---|
| 408 | """ |
|---|
| 409 | Output into topdl. Just call the topdl output, as the constraint |
|---|
| 410 | attributes should be in the topology. |
|---|
| 411 | """ |
|---|
| 412 | print >>f, topdl.topology_to_xml(comp, top='experiment') |
|---|
| 413 | |
|---|
| 414 | def ns2_out(f, top, constraints): |
|---|
| 415 | """ |
|---|
| 416 | Reformat the constraint data structures into ns2 constraint comments |
|---|
| 417 | and output the ns2 using the topdl routine. |
|---|
| 418 | """ |
|---|
| 419 | # Inner routines |
|---|
| 420 | # Deal with the possibility that the single string name is still in the |
|---|
| 421 | # constraint. |
|---|
| 422 | def name_format(n): |
|---|
| 423 | if isinstance(n, tuple): return n[0] |
|---|
| 424 | else: return n |
|---|
| 425 | |
|---|
| 426 | |
|---|
| 427 | # Format the required field back into a string. (ns2_out inner |
|---|
| 428 | def required_format(x): |
|---|
| 429 | if x: return "required" |
|---|
| 430 | else: return "optional" |
|---|
| 431 | |
|---|
| 432 | # ns2_out main line |
|---|
| 433 | for c in constraints: |
|---|
| 434 | print >>f, "# COMPOSITION: %s:%s:%s:%s" % ( |
|---|
| 435 | topdl.to_tcl_name(name_format(c.name)), |
|---|
| 436 | required_format(c.required), ",".join(c.provides), |
|---|
| 437 | ",".join(c.accepts)) |
|---|
| 438 | print >>f, topdl.topology_to_ns2(top) |
|---|
| 439 | |
|---|
| 440 | # Info to map from format to output routine. |
|---|
| 441 | exporters = { |
|---|
| 442 | 'xml':topdl_out, 'topdl':topdl_out, |
|---|
| 443 | 'tcl': ns2_out, 'ns': ns2_out, |
|---|
| 444 | } |
|---|
| 445 | |
|---|
| 446 | if format: |
|---|
| 447 | # Explicit format set, use it |
|---|
| 448 | if format in exporters: |
|---|
| 449 | exporter = exporters[format] |
|---|
| 450 | else: |
|---|
| 451 | raise RuntimeError("Unknown format %s" % format) |
|---|
| 452 | elif outfile: |
|---|
| 453 | # Determine the format from the suffix (if any) |
|---|
| 454 | s = get_suffix(outfile) |
|---|
| 455 | if s and s in exporters: |
|---|
| 456 | exporter = exporters[s] |
|---|
| 457 | else: |
|---|
| 458 | raise RuntimeError("Unknown format (suffix) %s" % outfile) |
|---|
| 459 | else: |
|---|
| 460 | # Both outfile and format are empty |
|---|
| 461 | exporter = topdl_out |
|---|
| 462 | |
|---|
| 463 | # The actual output. Open the file, if any, and call the exporter |
|---|
| 464 | if outfile: f = open(outfile, "w") |
|---|
| 465 | else: f = sys.stdout |
|---|
| 466 | exporter(f, top, constraints) |
|---|
| 467 | if outfile: f.close() |
|---|
| 468 | |
|---|
| 469 | def import_components(files): |
|---|
| 470 | """ |
|---|
| 471 | Pull in the components. The input is a sequence of tuples where each tuple |
|---|
| 472 | includes the name of the file to pull the component from and the number of |
|---|
| 473 | copies to make of it. The routine to read a file is picked by its suffix. |
|---|
| 474 | On completion, a tuple containing the number of components successfully |
|---|
| 475 | read, a list of the topologies, a set of names in use accross all |
|---|
| 476 | topologies (for inserting later names), the constraints extracted, and |
|---|
| 477 | indexes mapping the attribute provices and accepted tinto the lists of |
|---|
| 478 | constraints that provide or accept them is returned. |
|---|
| 479 | """ |
|---|
| 480 | importers = { |
|---|
| 481 | 'tcl': import_ns2_component, |
|---|
| 482 | 'ns': import_ns2_component, |
|---|
| 483 | 'xml':import_topdl_component, |
|---|
| 484 | 'topdl':import_topdl_component, |
|---|
| 485 | } |
|---|
| 486 | names = set() |
|---|
| 487 | constraints = [ ] |
|---|
| 488 | provides = { } |
|---|
| 489 | accepts = { } |
|---|
| 490 | components = 0 |
|---|
| 491 | topos = [ ] |
|---|
| 492 | |
|---|
| 493 | for fn, cnt in files: |
|---|
| 494 | top = None |
|---|
| 495 | try: |
|---|
| 496 | s = get_suffix(fn) |
|---|
| 497 | if s and s in importers: |
|---|
| 498 | top, marks = importers[s](fn) |
|---|
| 499 | else: |
|---|
| 500 | warn("Unknown suffix on file %s. Ignored" % fn) |
|---|
| 501 | continue |
|---|
| 502 | except service_error, e: |
|---|
| 503 | warn("Remote error on %s: %s" % (fn, e)) |
|---|
| 504 | continue |
|---|
| 505 | except EnvironmentError, e: |
|---|
| 506 | warn("Error on %s: %s" % (fn, e)) |
|---|
| 507 | continue |
|---|
| 508 | |
|---|
| 509 | # Parsed the component and sonctraints, now work through the rest of |
|---|
| 510 | # the pre-processing. We do this once per copy of the component |
|---|
| 511 | # requested, cloning topologies and constraints each time. |
|---|
| 512 | for i in range(0, cnt): |
|---|
| 513 | components += 1 |
|---|
| 514 | t = top.clone() |
|---|
| 515 | m = copy.deepcopy(marks) |
|---|
| 516 | index_constraints(m.values(), provides, accepts) |
|---|
| 517 | add_interfaces(t, m) |
|---|
| 518 | localize_names(t, names, m) |
|---|
| 519 | t.incorporate_elements() |
|---|
| 520 | constraints.extend(m.values()) |
|---|
| 521 | topos.append(t) |
|---|
| 522 | |
|---|
| 523 | return (components, topos, names, constraints, provides, accepts) |
|---|
| 524 | |
|---|
| 525 | # Main line begins |
|---|
| 526 | parser = ComposeOptionParser() |
|---|
| 527 | |
|---|
| 528 | opts, args = parser.parse_args() |
|---|
| 529 | |
|---|
| 530 | if opts.cert: |
|---|
| 531 | cert = opts.cert |
|---|
| 532 | elif os.access(os.path.expanduser("~/.ssl/emulab.pem"), os.R_OK): |
|---|
| 533 | cert = os.path.expanduser("~/.ssl/emulab.pem") |
|---|
| 534 | else: |
|---|
| 535 | cert = None |
|---|
| 536 | |
|---|
| 537 | random.seed(opts.seed) |
|---|
| 538 | |
|---|
| 539 | files = opts.files |
|---|
| 540 | files.extend([ (a, 1) for a in args]) |
|---|
| 541 | |
|---|
| 542 | # Pull 'em in. |
|---|
| 543 | components, topos, names, constraints, provides, accepts = \ |
|---|
| 544 | import_components(files) |
|---|
| 545 | |
|---|
| 546 | # Let the user know if they messed up on specifying constraints. |
|---|
| 547 | if any([ not isinstance(c.name, tuple) for c in constraints]): |
|---|
| 548 | warn("nodes not found for constraints on %s" % \ |
|---|
| 549 | ",".join([ c.name for c in constraints \ |
|---|
| 550 | if isinstance(c.name, basestring)])) |
|---|
| 551 | constraints = [ c for c in constraints if isinstance(c.name, tuple )] |
|---|
| 552 | |
|---|
| 553 | # If more than one component was given, actually do the composition, otherwise |
|---|
| 554 | # this is probably a format conversion. |
|---|
| 555 | if components > 1: |
|---|
| 556 | # Mix up the constraint indexes |
|---|
| 557 | randomize_constraint_order((provides, accepts)) |
|---|
| 558 | |
|---|
| 559 | # Now the various components live in the same namespace and are marked with |
|---|
| 560 | # their composition requirements. |
|---|
| 561 | |
|---|
| 562 | if not meet_constraints([c for c in constraints if c.required], |
|---|
| 563 | provides, accepts): |
|---|
| 564 | if opts.lax: |
|---|
| 565 | warn("warning: could not meet all required constraints") |
|---|
| 566 | else: |
|---|
| 567 | sys.exit("Failed: could not meet all required constraints") |
|---|
| 568 | |
|---|
| 569 | meet_constraints([ c for c in constraints if not c.match ], |
|---|
| 570 | provides, accepts) |
|---|
| 571 | |
|---|
| 572 | # Add testbed attributes if requested |
|---|
| 573 | if opts.add_testbeds: |
|---|
| 574 | for i, t in enumerate(topos): |
|---|
| 575 | for e in [ e for e in t.elements if isinstance(e, topdl.Computer)]: |
|---|
| 576 | e.set_attribute('testbed', 'testbed%03d' % i) |
|---|
| 577 | |
|---|
| 578 | # Make a topology containing all elements and substrates from components |
|---|
| 579 | # that had matches. |
|---|
| 580 | comp = topdl.Topology() |
|---|
| 581 | for t in set([ c.name[2] for c in constraints if c.match]): |
|---|
| 582 | comp.elements.extend([e.clone() for e in t.elements]) |
|---|
| 583 | comp.substrates.extend([s.clone() for s in t.substrates]) |
|---|
| 584 | |
|---|
| 585 | # Add substrates and connections corresponding to composition points |
|---|
| 586 | connect_composition_points(comp, constraints, names) |
|---|
| 587 | |
|---|
| 588 | # Remove composition attributes for satisfied constraints |
|---|
| 589 | remove_constraint_attributes(comp, dict( |
|---|
| 590 | [(c.name[0], c) for c in constraints if c.match])) |
|---|
| 591 | elif topos == 1: |
|---|
| 592 | comp = topos[0] |
|---|
| 593 | if opts.add_testbeds: |
|---|
| 594 | for e in [ e for e in comp.elements if isinstance(e, topdl.Computer)]: |
|---|
| 595 | e.set_attribute('testbed', 'testbed001') |
|---|
| 596 | |
|---|
| 597 | else: |
|---|
| 598 | sys.exit("Did not read any components.") |
|---|
| 599 | |
|---|
| 600 | # Put out the composition with only the unmatched constriaints |
|---|
| 601 | output_composition(comp, [c for c in constraints if not c.match], |
|---|
| 602 | opts.outfile, opts.format) |
|---|